digitaldna.LongestCommonSubsequence¶
-
class
digitaldna.LongestCommonSubsequence(in_path='', out_path='/tmp/glcr_cache', overwrite=False, threshold='auto', window=10, verbosity=2)[source]¶ The Digital DNA Python implementation.
Parameters: - in_path : str, optional
The name with absolute path of a file containing the sequences you want to process. The input file must be a txt file and the first row must contain the number of sequences to read. Default: ‘’
- out_path : str, optional
The output file name with absolute path of the file where the algorithm will save result in case of verbosity equals to Verbosity.FILE or Verbosity.FILE_EXTENDED. Default: ‘/tmp/glcr_cache’
- overwrite : boolean, optional
It must be False to use the LCS files produced in a previous fit call, in this case the file names are the ones specified in the out_path parameter. If True, recomputes the LCS files. Default: False
- threshold : boolean, optional
It must be False to use the LCS files produced in a previous fit call, in this case the file names are the ones specified in the out_path parameter. If True, recomputes the LCS files. Default: False
- window : str, optional
The size of the window used to compute the cutting threshold between bot and not bot. The cutting point is computed by smoothing the curve, deriving the result and taking the first (l.h.s.) local maxima. The window parameter influences both smoothing and finding the local maxima. It must be 2 < window < n_accounts. Default: 10
- verbosity : str, optional
The verbosity parameter is used to specify whether to save results to files or not. It must be:
TEST does not write anything, used for benchmarking
MEMORY_ONLY retrieves only the couples (sequence length, # of accounts), used for plots
FILE produces 2 files, a file named out_path + ‘.gsa’ where each row contains the identifier of the sequence. In the other file, named out_path + ‘.mat’, each row contains:
- sequence length
- # of accounts
- range of indexes (begin and end)
FILE_EXTENDED as FILE but the in_path + ‘.mat’ file contains also the column of the common subsequence
References
S. Cresci, R. D. Pietro, M. Petrocchi, A. Spognardi and M. Tesconi, “Social Fingerprinting: Detection of Spambot Groups Through DNA-Inspired Behavioral Modeling”, IEEE Transactions on Dependable and Secure Computing, vol. 15, no. 4, pp. 561-576, 1 July-Aug. 2018, https://ieeexplore.ieee.org/document/7876716
S. Cresci, R. di Pietro, M. Petrocchi, A. Spognardi and M. Tesconi, “Exploiting Digital DNA for the Analysis of Similarities in Twitter Behaviours”, 2017 IEEE International Conference on Data Science and Advanced Analytics (DSAA), Tokyo, 2017, pp. 686-695, https://ieeexplore.ieee.org/document/8259831
M. Arnold, E. Ohlebusch, “Linear Time Algorithms for Generalizations of the Longest Common Substring Problem”, Algorithmica, vol 60, pp. 806-818, 4 August 2011, https://link.springer.com/article/10.1007/s00453-009-9369-1
Methods
fit(X[, y])Computes the longest common subsequence fit_predict(X[, y])“Fits the model to the training set X and returns the labels (True for bot, False for non bot) on the training set according to the window parameter. get_params([deep])Get parameters for this estimator. plot_LCS()“Plots the longest common subsequence curve as (number of accounts, sequence length) plot_LCS_log()“Plots the longest common subsequence curve as (log(number of accounts), log(sequence length)) predict([X])Predict the labels (True bot, False Not Bot) of X according to lcs and window parameter:. set_params(**params)Set the parameters of this estimator. -
__init__(in_path='', out_path='/tmp/glcr_cache', overwrite=False, threshold='auto', window=10, verbosity=2)[source]¶ Initialize self. See help(type(self)) for accurate signature.
-
fit(X, y=None)[source]¶ Computes the longest common subsequence
Parameters: - X : array-like or sparse matrix of shape = [n_samples, n_features]
The training input samples.
- y : None
There is no need of a target in a transformer, yet the pipeline API requires this parameter.
Returns: - self : object
Returns self.
Attributes: - lcs_index_ : pandas dataframe, shape (distinct couples (lcs, n_of_accounts), 2), default=None
The dataframe containing the distict couples lcs, n_of_accounts). Only if: verbosity == Verbosity.MEMORY_ONLY
-
fit_predict(X, y=None)[source]¶ “Fits the model to the training set X and returns the labels (True for bot, False for non bot) on the training set according to the window parameter. Parameters ———- X : array-like, shape (n_samples, n_features), default=None
The query sample or samples to compute the Local Outlier Factor w.r.t. to the training samples.Returns: - y : array, shape (n_samples,)
Returns True for bots and False for real timeline.
Attributes: - lcs_index_ : pandas dataframe, shape (distinct couples (lcs, n_of_accounts), 2), default=None
The dataframe containing the distict couples lcs, n_of_accounts)
-
get_params(deep=True)¶ Get parameters for this estimator.
Parameters: - deep : boolean, optional
If True, will return the parameters for this estimator and contained subobjects that are estimators.
Returns: - params : mapping of string to any
Parameter names mapped to their values.
-
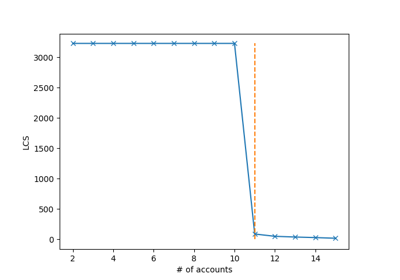
plot_LCS()[source]¶ “Plots the longest common subsequence curve as (number of accounts, sequence length)
Returns: - self : returns an instance of self.
Attributes: - lcs_index_ : pandas dataframe, shape (distinct couples (lcs, n_of_accounts), 2), default=None
The dataframe containing the distict couples lcs, n_of_accounts)
-
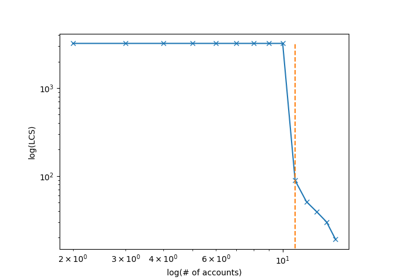
plot_LCS_log()[source]¶ “Plots the longest common subsequence curve as (log(number of accounts), log(sequence length))
Returns: - self : returns an instance of self.
Attributes: - lcs_index_ : pandas dataframe, shape (distinct couples (lcs, n_of_accounts), 2), default=None
The dataframe containing the distict couples lcs, n_of_accounts)
-
predict(X=None)[source]¶ Predict the labels (True bot, False Not Bot) of X according to lcs and window parameter:. If X is None, returns the same as fit_predict(X_train). Parameters ———- X : array-like, shape (n_samples, n_features), default=None
The query sample or samples to identify bot groups. If None, makes prediction on the training data.- y : array, shape (n_samples,)
- Returns True for bots and False for real timeline.
-
set_params(**params)¶ Set the parameters of this estimator.
The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.Returns: - self