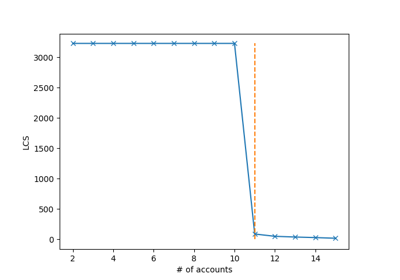
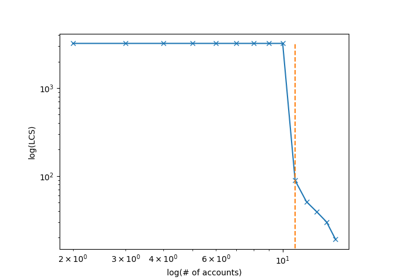
digitaldna.TwitterDDNASequencer¶
-
class
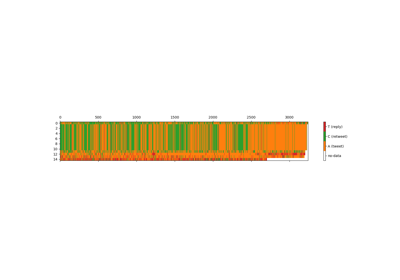
digitaldna.TwitterDDNASequencer(alphabet='b3_type', input_file='')[source]¶ Twitter Digital DNA Sequencer. Compute sequences of digital DNA from twitter timelines (check out https://developer.twitter.com/en/docs/tweets/timelines/api-reference/get-statuses-user_timeline.html)
Parameters: - alphabet : string or callable, default ‘b3_type’
mapping between the column value and the corresponding base. If alphabet_ is a callable function, it is called on each pair of instances (rows) and the resulting value recorded. The callable should take two arrays as input and return one value indicating the distance between them. Prebuild alphabets are the following:
- ‘b3_type’, where the correspondence is
- ‘A’ for tweet
- ‘C’ for reply
- ‘T’ for retweet
- ‘b3_content’, where the correspondence is
- ‘N’ tweet contains no entities (plain text)
- ‘E’ tweet contains entities of one type
- ‘X’ tweet contains entities of mixed types
- ‘b6_content’, where the correspondence is
- ‘N’ tweet contains no entities (plain text)
- ‘U’ tweet contains one or more URLs
- ‘H’ tweet contains one or more hashtags
- ‘M’ tweet contains one or more mentions
- ‘D’ tweet contains one or more medias
- ‘X’ tweet contains entities of mixed types
References
S. Cresci, R. D. Pietro, M. Petrocchi, A. Spognardi and M. Tesconi, “Social Fingerprinting: Detection of Spambot Groups Through DNA-Inspired Behavioral Modeling”, IEEE Transactions on Dependable and Secure Computing, vol. 15, no. 4, pp. 561-576, 1 July-Aug. 2018, https://ieeexplore.ieee.org/document/7876716
S. Cresci, R. di Pietro, M. Petrocchi, A. Spognardi and M. Tesconi, “Exploiting Digital DNA for the Analysis of Similarities in Twitter Behaviours”, 2017 IEEE International Conference on Data Science and Advanced Analytics (DSAA), Tokyo, 2017, pp. 686-695, https://ieeexplore.ieee.org/document/8259831
Attributes: - input_shape : tuple
The shape the data passed to
fit()
Methods
fit([X, y])Simply assigns the right alphabet mapper function to remap_ fit_transform([X, y])Fit and transform get_params()Get parameters for this estimator. set_params(alphabet, input_file)Set the parameters of this estimator. transform([X])The function that transform the array of timelines to digital dna sequences given the alphabet. -
__init__(alphabet='b3_type', input_file='')[source]¶ Initialize self. See help(type(self)) for accurate signature.
-
fit(X=None, y=None)[source]¶ Simply assigns the right alphabet mapper function to remap_
Parameters: - X : None
The pipeline API requires this parameter.
- y : None
There is no need of a target in a transformer, yet the pipeline API requires this parameter.
Returns: - self : object
Returns an instance of self.
Attributes: - remap_ : function that takes a tweet with the same format retrieved from
GET statuses/user_timeline call and retrieves the corresponding character given the alphabet parameter.
-
fit_transform(X=None, y=None)[source]¶ Fit and transform
Parameters: - X : array-like of shape = [# of tweets, 1]
The input samples, each sample is a python dict of a tweet as retrieved from twitter user timelines API (check out https://developer.twitter.com/en/docs/tweets/timelines/api-reference/get-statuses-user_timeline.html)
- y: ignored parameter needed to mantain a standard pattern
Returns: - X_transformed : array of shape = [n_samples, 2]
The resulting array where the first column is the user id and the second is the translated sequence
-
get_params()[source]¶ Get parameters for this estimator.
Returns: - params : mapping of string to any
Parameter names mapped to their values.
-
set_params(alphabet, input_file)[source]¶ Set the parameters of this estimator. The method works on simple estimators as well as on nested objects (such as pipelines). The latter have parameters of the form
<component>__<parameter>so that it’s possible to update each component of a nested object.Returns: - self
-
transform(X=None)[source]¶ The function that transform the array of timelines to digital dna sequences given the alphabet.
Parameters: - X : array-like of shape = [# of tweets, 1]
The input samples, each sample is a python dict of a tweet as retrieved from twitter user timelines API (check out https://developer.twitter.com/en/docs/tweets/timelines/api-reference/get-statuses-user_timeline.html)
Returns: - X_transformed : array of string of shape = [# of users, 1]
The array containing the digital dna sequences